ASL Protocols
Please contact cfmri@ucsd.edu for any questions about this page.
ASL Protocols Overview
CFMRI supports the following standard protocols on the Siemens Prisma scanner. The 3mm protocol is our standard recommendation as it provides a good blend of SNR and image resolution for most imaging studies. All protocols combine a PCASL preparation with a 3D GRASE readout, along with excellent whole-brain coverage.
On the scanner, these protocols are located under CFMRI_Templates > Arterial Spin Labeling > Cerebral Blood Flow > 3D PCASL.
These protocols should be used with either the 32ch or 64ch head coils. If you must use the 20ch head coil with ASL, please contact us for further discussion.
Protocol |
Low-Res |
3mm |
2.5mm |
|---|---|---|---|
Voxel Size |
3.75mm x 3.75mm x 4mm |
3mm isotropic |
2.5mm isotropic |
Matrix Size |
64 x 64 x 36 |
74 x 74 x 50 |
90 x 90 x 50 |
Field of View (FOV) |
240mm x 240mm x 144mm |
222mm x 222mm x 150mm |
225mm x 225mm x 150mm |
TE |
12.94ms |
15.42ms |
18.1ms |
TR |
4000ms |
4100ms |
4200ms |
M0 TR |
5000ms |
5000ms |
5000ms |
Flip Angle |
120 |
120 |
120 |
Labeling Duration |
1800ms |
1800ms |
1800ms |
Post-Labeling Delay |
2000ms |
2000ms |
2000ms |
Tag Gap |
20mm |
20mm |
20mm |
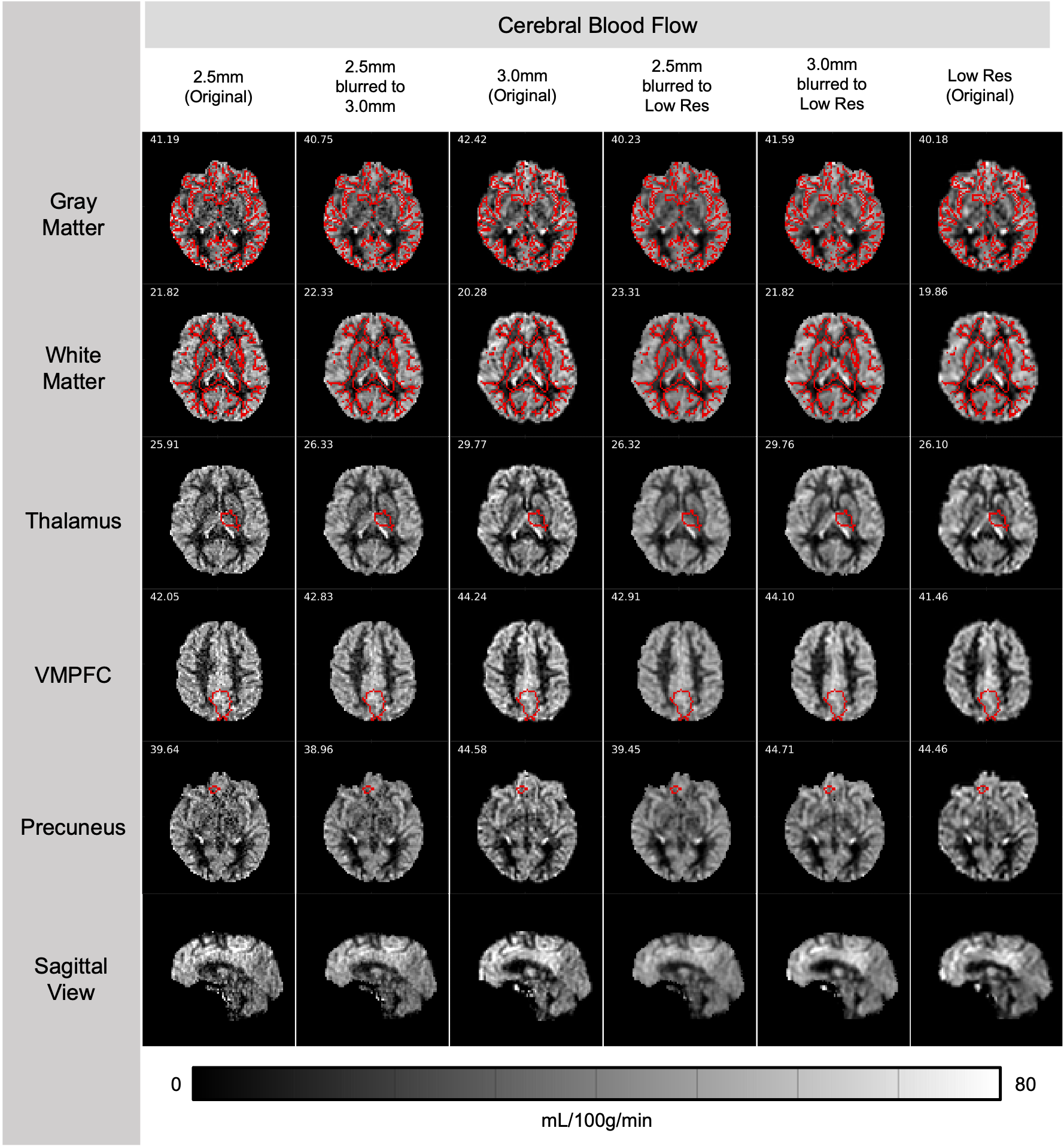
Sample Comparison
Below is a comparison of CBF maps from the three protocols collected on a healthy 27-y.o. participant. The numbers indicate the median CBF value within the ROI outlined in red. All scans were ~7 minutes long with 24 tag/control pairs collected.
Sequence Structure
Each TR consists of a pre-saturation pulse, PCASL labeling, a post-labeling delay, and finally the image readout. Interspersed through the PCASL labeling and post-labeling delay period are four background suppression (BGS) pulses, timed to achieve approximately 95% suppression of static tissue. With the combination of PCASL labeling and the four BGS pulses, the overall labeling efficiency is about 0.6.
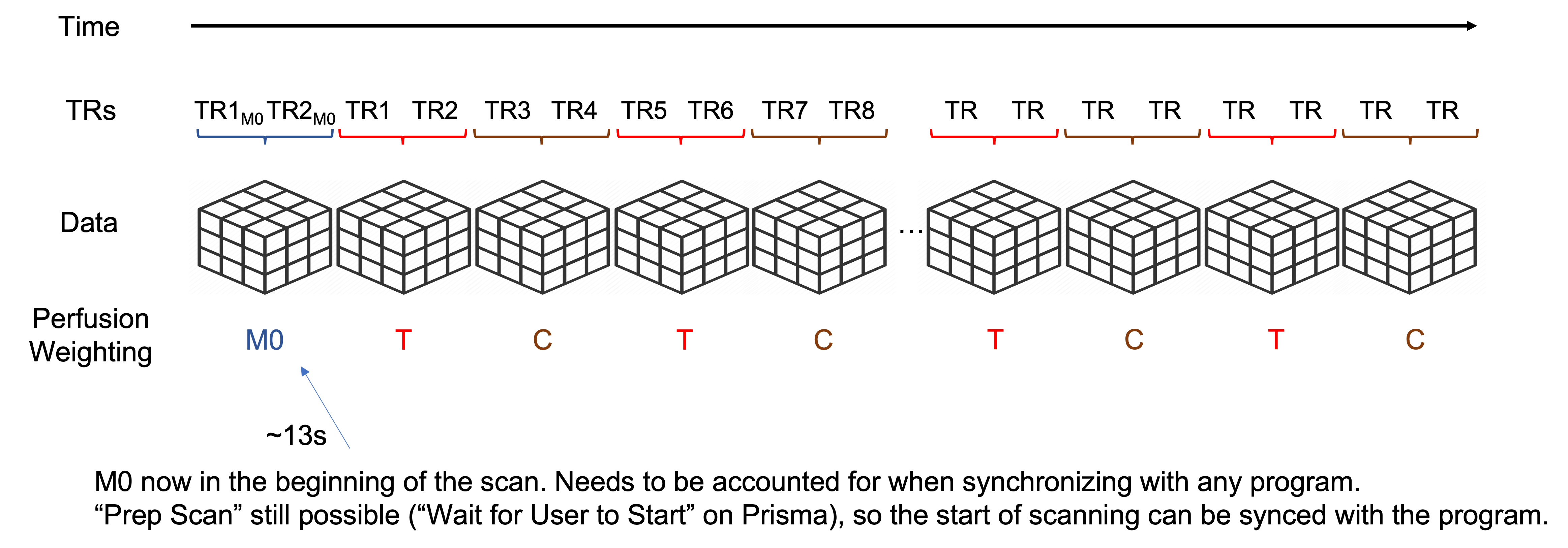
Scan Structure
The scan data consists of an initial M0 scan followed by alternating tag/control/tag/control/… volumes until the end of the scan. These protocols require 2 segments per volume, meaning it requires two TRs worth of data to construct one image volume.
The initial M0 image is acquired with a TR of 5000ms. This provides enough time for approximately full tissue relaxation to achieve a proton-density weighted scan contrast. The M0 and the initial reference scanning together comprise the initial ~13s of the scan. After that begins the first TR for the first tag image. If you are performing a task alongside the ASL scan, please account for the initial 13s of scanning. You can either manually synchronize your task with the start of the first tag TR, or trigger your task with the first M0 TR and build in a 13s buffer to your task.
A diagram of the scan structure is given below.